
Back Molekuuldinamieksimulasie Afrikaans ديناميكا جزيئية Arabic Dinàmica molecular Catalan Molekulardynamik-Simulation German Dinámica molecular Spanish Molekulaardünaamika Estonian دینامیک ملکولی Persian Molekyylidynamiikka Finnish Dynamique moléculaire French דינמיקה מולקולרית HE

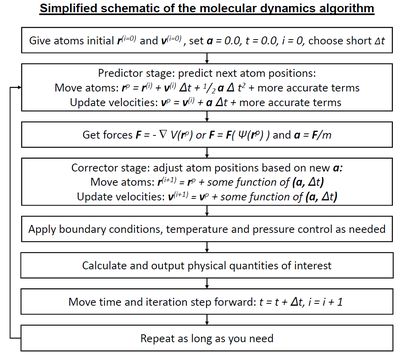
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics.
Because molecular systems typically consist of a vast number of particles, it is impossible to determine the properties of such complex systems analytically; MD simulation circumvents this problem by using numerical methods. However, long MD simulations are mathematically ill-conditioned, generating cumulative errors in numerical integration that can be minimized with proper selection of algorithms and parameters, but not eliminated.
For systems that obey the ergodic hypothesis, the evolution of one molecular dynamics simulation may be used to determine the macroscopic thermodynamic properties of the system: the time averages of an ergodic system correspond to microcanonical ensemble averages. MD has also been termed "statistical mechanics by numbers" and "Laplace's vision of Newtonian mechanics" of predicting the future by animating nature's forces[1] and allowing insight into molecular motion on an atomic scale.
- ^ Schlick T (1996). "Pursuing Laplace's Vision on Modern Computers". Mathematical Approaches to Biomolecular Structure and Dynamics. The IMA Volumes in Mathematics and its Applications. Vol. 82. pp. 219–247. doi:10.1007/978-1-4612-4066-2_13. ISBN 978-0-387-94838-6.
© MMXXIII Rich X Search. We shall prevail. All rights reserved. Rich X Search

