
Back سكيراء الجعة Arabic سكيراء الجعه ARZ Fisijski kvasac BS Schizosaccharomyces pombe CEB Schizosaccharomyces pombe German Schizosaccharomyces pombe Spanish Schizosaccharomyces pombe Basque Schizosaccharomyces pombe French Schizosaccharomyces pombe Galician שמר החלוקה HE
| Schizosaccharomyces pombe | |
|---|---|
| |
| Scientific classification | |
| Domain: | Eukaryota |
| Kingdom: | Fungi |
| Division: | Ascomycota |
| Class: | Schizosaccharomycetes |
| Order: | Schizosaccharomycetales |
| Family: | Schizosaccharomycetaceae |
| Genus: | Schizosaccharomyces |
| Species: | S. pombe
|
| Binomial name | |
| Schizosaccharomyces pombe Lindner (1893)
| |
| Synonyms[1] | |
| |
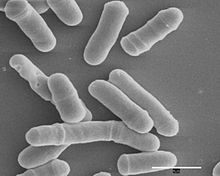
Schizosaccharomyces pombe, also called "fission yeast", is a species of yeast used in traditional brewing and as a model organism in molecular and cell biology. It is a unicellular eukaryote, whose cells are rod-shaped. Cells typically measure 3 to 4 micrometres in diameter and 7 to 14 micrometres in length. Its genome, which is approximately 14.1 million base pairs, is estimated to contain 4,970 protein-coding genes and at least 450 non-coding RNAs.[2]
These cells maintain their shape by growing exclusively through the cell tips and divide by medial fission to produce two daughter cells of equal size, which makes them a powerful tool in cell cycle research.
Fission yeast was isolated in 1893 by Paul Lindner from East African millet beer. The species name pombe is the Swahili word for beer. It was first developed as an experimental model in the 1950s: by Urs Leupold for studying genetics,[3][4] and by Murdoch Mitchison for studying the cell cycle.[5][6][7]
Paul Nurse, a fission yeast researcher, successfully merged the independent schools of fission yeast genetics and cell cycle research. Together with Lee Hartwell and Tim Hunt, Nurse won the 2001 Nobel Prize in Physiology or Medicine for work on cell cycle regulation.
The sequence of the S. pombe genome was published in 2002, by a consortium led by the Sanger Institute, becoming the sixth model eukaryotic organism whose genome has been fully sequenced. S. pombe researchers are supported by the PomBase MOD (model organism database). This has fully unlocked the power of this organism, with many genes orthologous to human genes identified - 70% to date,[8][9] including many genes involved in human disease.[10] In 2006, sub-cellular localization of almost all the proteins in S. pombe was published using green fluorescent protein as a molecular tag.[11]
Schizosaccharomyces pombe has also become an important organism in studying the cellular responses to DNA damage and the process of DNA replication.
Approximately 160 natural strains of S. pombe have been isolated. These have been collected from a variety of locations including Europe, North and South America, and Asia. The majority of these strains have been collected from cultivated fruits such as apples and grapes, or from the various alcoholic beverages, such as Brazilian Cachaça. S. pombe is also known to be present in fermented tea, kombucha.[12] It is not clear at present whether S. pombe is the major fermenter or a contaminant in such brews. The natural ecology of Schizosaccharomyces yeasts is not well-studied.
- ^ "Schizosaccharomyces pombe". Global Biodiversity Information Facility. Retrieved 14 August 2021.
- ^ Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, et al. (June 2008). "Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution". Nature. 453 (7199): 1239–43. Bibcode:2008Natur.453.1239W. doi:10.1038/nature07002. PMID 18488015. S2CID 205213499.
- ^ Leupold U (1950). "Die Vererbung von Homothallie und Heterothallie bei Schizosaccharomyces pombe". C R Trav Lab Carlsberg Ser Physiol. 24: 381–480.
- ^ Leupold U. (1993) The origins of Schizosaccharomyces pombe genetics. In: Hall MN, Linder P. eds. The Early Days of Yeast Genetics. New York. Cold Spring Harbor Laboratory Press. p 125–128.
- ^ Mitchison JM (October 1957). "The growth of single cells. I. Schizosaccharomyces pombe". Experimental Cell Research. 13 (2): 244–62. doi:10.1016/0014-4827(57)90005-8. PMID 13480293.
- ^ Mitchison JM (April 1990). "The fission yeast, Schizosaccharomyces pombe". BioEssays. 12 (4): 189–91. doi:10.1002/bies.950120409. PMID 2185750.
- ^ Fantes PA, Hoffman CS (June 2016). "A Brief History of Schizosaccharomyces pombe Research: A Perspective Over the Past 70 Years". Genetics. 203 (2): 621–9. doi:10.1534/genetics.116.189407. PMC 4896181. PMID 27270696.
- ^ Cite error: The named reference
pmid22039153was invoked but never defined (see the help page). - ^ "PomBase".
- ^ "PomBase".
- ^ Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, et al. (July 2006). "ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe". Nature Biotechnology. 24 (7): 841–7. doi:10.1038/nbt1222. PMID 16823372. S2CID 10397608.
- ^ Teoh AL, Heard G, Cox J (September 2004). "Yeast ecology of Kombucha fermentation". International Journal of Food Microbiology. 95 (2): 119–26. doi:10.1016/j.ijfoodmicro.2003.12.020. PMID 15282124.
© MMXXIII Rich X Search. We shall prevail. All rights reserved. Rich X Search