
Back معزز Arabic Amplificador genètic Catalan Enhancer Czech Enhancer (Genetik) German Ενισχυτής (γενετική) Greek Enhancer Spanish افزاینده Persian Amplificateur (biologie) French Feabhsaitheoir (bitheolaíocht) Irish Amplificador xenético Galician

- DNA
- Enhancer
- Promoter
- Gene
- Transcription Activator Protein
- Mediator Protein
- RNA Polymerase
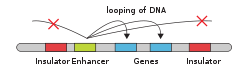
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins (activators) to increase the likelihood that transcription of a particular gene will occur.[1][2] These proteins are usually referred to as transcription factors. Enhancers are cis-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site.[2][3] There are hundreds of thousands of enhancers in the human genome.[2] They are found in both prokaryotes and eukaryotes.[4] Active enhancers typically get transcribed as enhancer or regulatory non-coding RNA, whose expression levels correlate with mRNA levels of target genes.[5]
The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983.[6][7][8] This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive.[9] Lately, enhancers have been shown to be involved in certain medical conditions, for example, myelosuppression.[10] Since 2022, scientists have used artificial intelligence to design synthetic enhancers and applied them in animal systems, first in a cell line,[11] and one year later also in vivo.[12][13]
- ^ Blackwood EM, Kadonaga JT (July 1998). "Going the distance: a current view of enhancer action". Science. 281 (5373): 60–63. Bibcode:1998Sci...281...60.. doi:10.1126/science.281.5373.60. PMID 9679020. S2CID 11666739.
- ^ a b c Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G (April 2013). "Enhancers: five essential questions". Nature Reviews. Genetics. 14 (4): 288–295. doi:10.1038/nrg3458. PMC 4445073. PMID 23503198.
- ^ Maston GA, Evans SK, Green MR (2006). "Transcriptional regulatory elements in the human genome". Annual Review of Genomics and Human Genetics. 7: 29–59. doi:10.1146/annurev.genom.7.080505.115623. PMID 16719718. S2CID 12346247.
- ^ Kulaeva OI, Nizovtseva EV, Polikanov YS, Ulianov SV, Studitsky VM (December 2012). "Distant activation of transcription: mechanisms of enhancer action". Molecular and Cellular Biology. 32 (24): 4892–4897. doi:10.1128/MCB.01127-12. PMC 3510544. PMID 23045397.
- ^ Burren OS, Rubio García A, Javierre BM, Rainbow DB, Cairns J, Cooper NJ, et al. (4 September 2017). "Chromosome contacts in activated T cells identify autoimmune disease candidate genes". Genome Biology. 18 (1): 165. doi:10.1186/s13059-017-1285-0. ISSN 1474-760X. PMC 5584004. PMID 28870212.
- ^ Mercola M, Wang XF, Olsen J, Calame K (August 1983). "Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus". Science. 221 (4611): 663–665. Bibcode:1983Sci...221..663M. doi:10.1126/science.6306772. PMID 6306772.
- ^ Banerji J, Olson L, Schaffner W (July 1983). "A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes". Cell. 33 (3): 729–740. doi:10.1016/0092-8674(83)90015-6. PMID 6409418. S2CID 23981549.
- ^ Gillies SD, Morrison SL, Oi VT, Tonegawa S (July 1983). "A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene". Cell. 33 (3): 717–728. doi:10.1016/0092-8674(83)90014-4. PMID 6409417. S2CID 40313833.
- ^ Hauptman G, Reichert MC, Abdal Rhida MA, Evans TA (December 2022). "Characterization of enhancer fragments in Drosophila robo2". Fly. 16 (1): 312–346. bioRxiv 10.1101/2022.08.01.502399. doi:10.1080/19336934.2022.2126259. PMC 9559326. PMID 36217698.
- ^ Zhigulev A, Norberg Z, Cordier J, Spalinskas R, Bassereh H, Björn N, et al. (March 2024). "Enhancer mutations modulate the severity of chemotherapy-induced myelosuppression". Life Science Alliance. 7 (3): e202302244. doi:10.26508/lsa.202302244. PMC 10796589. PMID 38228368.
- ^ de Almeida BP, Reiter F, Pagani M, Stark A (May 2022). "DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers". Nature Genetics. 54 (5): 613–624. doi:10.1038/s41588-022-01048-5. PMID 35551305.
- ^ de Almeida BP, Schaub C, Pagani M, Secchia S, Furlong EE, Stark A (February 2024). "Targeted design of synthetic enhancers for selected tissues in the Drosophila embryo". Nature. 626 (7997): 207–211. Bibcode:2024Natur.626..207D. doi:10.1038/s41586-023-06905-9. PMC 10830412. PMID 38086418.
- ^ Taskiran II, Spanier KI, Dickmänken H, Kempynck N, Pančíková A, Ekşi EC, et al. (February 2024). "Cell-type-directed design of synthetic enhancers". Nature. 626 (7997): 212–220. Bibcode:2024Natur.626..212T. doi:10.1038/s41586-023-06936-2. PMC 10830415. PMID 38086419.
© MMXXIII Rich X Search. We shall prevail. All rights reserved. Rich X Search