
Back مجهريات البقعة Arabic Mikrobiota BS Microbiota Catalan Mikrobioto Esperanto Microbiota Spanish Mikrobioota Estonian میکروبیوتا Persian Microbiote French מיקרוביוטה HE Mikrobiota ID
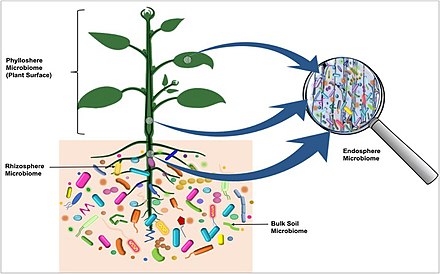
Microbiota are the range of microorganisms that may be commensal, mutualistic, or pathogenic found in and on all multicellular organisms, including plants. Microbiota include bacteria, archaea, protists, fungi, and viruses,[2][3] and have been found to be crucial for immunologic, hormonal, and metabolic homeostasis of their host.
The term microbiome describes either the collective genomes of the microbes that reside in an ecological niche or else the microbes themselves.[4][5][6]
The microbiome and host emerged during evolution as a synergistic unit from epigenetics and genetic characteristics, sometimes collectively referred to as a holobiont.[7][8] The presence of microbiota in human and other metazoan guts has been critical for understanding the co-evolution between metazoans and bacteria.[9][10] Microbiota play key roles in the intestinal immune and metabolic responses via their fermentation product (short-chain fatty acid), acetate.[11]
- ^ Dastogeer, K.M., Tumpa, F.H., Sultana, A., Akter, M.A. and Chakraborty, A. (2020) "Plant microbiome–an account of the factors that shape community composition and diversity". Current Plant Biology, 23: 100161. doi:10.1016/j.cpb.2020.100161.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ De Sordi, Luisa; Lourenço, Marta; Debarbieux, Laurent (2019). "The battle within: interactions of bacteriophages and bacteria in the gastrointestinal tract". Cell Host & Microbe. 25 (2): 210–218. doi:10.1016/j.chom.2019.01.018. PMID 30763535. S2CID 73455329.
- ^ Peterson, J; Garges, S; et al. (2009). "The NIH Human Microbiome Project". Genome Research. 19 (12). NIH HMP Working Group: 2317–2323. doi:10.1101/gr.096651.109. PMC 2792171. PMID 19819907.
- ^ Backhed, F.; Ley, R. E.; Sonnenburg, J. L.; Peterson, D. A.; Gordon, J. I. (2005). "Host-Bacterial Mutualism in the Human Intestine". Science. 307 (5717): 1915–1920. Bibcode:2005Sci...307.1915B. doi:10.1126/science.1104816. PMID 15790844. S2CID 6332272.
- ^ Turnbaugh, P. J.; Ley, R. E.; Hamady, M.; Fraser-Liggett, C. M.; Knight, R.; Gordon, J. I. (2007). "The Human Microbiome Project". Nature. 449 (7164): 804–810. Bibcode:2007Natur.449..804T. doi:10.1038/nature06244. PMC 3709439. PMID 17943116.
- ^ Ley, R. E.; Peterson, D. A.; Gordon, J. I. (2006). "Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine". Cell. 124 (4): 837–848. doi:10.1016/j.cell.2006.02.017. PMID 16497592. S2CID 17203181.
- ^ Salvucci, E. (2016). "Microbiome, holobiont and the net of life". Critical Reviews in Microbiology. 42 (3): 485–494. doi:10.3109/1040841X.2014.962478. hdl:11336/33456. PMID 25430522. S2CID 30677140.
- ^ Guerrero, R.; Margulis, Lynn; Berlanga, M. (2013). "Symbiogenesis: The holobiont as a unit of evolution". International Microbiology. 16 (3): 133–143. doi:10.2436/20.1501.01.188. PMID 24568029.
- ^ Davenport, Emily R et al. "The human microbiome in evolution". BMC Biology. vol. 15,1 127. 27 Dec. 2017, doi:10.1186/s12915-017-0454-7
- ^ "Evolution of the human gut flora". Andrew H. Moeller, Yingying Li, Eitel Mpoudi Ngole, Steve Ahuka-Mundeke, Elizabeth V. Lonsdorf, Anne E. Pusey, Martine Peeters, Beatrice H. Hahn, Howard Ochman. Proceedings of the National Academy of Sciences. Nov 2014, 111 (46) 16431–16435; doi:10.1073/pnas.1419136111
- ^ Jugder, Bat-Erdene; Kamareddine, Layla; Watnick, Paula I. (2021). "Microbiota-derived acetate activates intestinal innate immunity via the Tip60 histone acetyltransferase complex". Immunity. 54 (8): 1683–1697.e3. doi:10.1016/j.immuni.2021.05.017. ISSN 1074-7613. PMC 8363570. PMID 34107298.
© MMXXIII Rich X Search. We shall prevail. All rights reserved. Rich X Search