
Back Fotolijaza BS Fotoliasa Catalan Photolyasen German Fotoliasa Spanish Photolyase French Fotoliase Galician Deossiribodipirimidina foto-liasi Italian フォトリアーゼ Japanese Fotoliaza Polish Fotoliase Portuguese
| Cryptochrome/photolyase, C-terminal, FAD binding | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
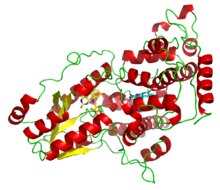 A deazaflavin photolyase from Anacystis nidulans, illustrating the two light-harvesting cofactors: FADH− (yellow) and 8-HDF (cyan). | |||||||||||
| Identifiers | |||||||||||
| Symbol | FAD_binding_7 | ||||||||||
| Pfam | PF03441 | ||||||||||
| InterPro | IPR005101 | ||||||||||
| PROSITE | PDOC00331 | ||||||||||
| SCOP2 | 1qnf / SCOPe / SUPFAM | ||||||||||
| |||||||||||
| deoxyribodipyrimidine photo-lyase (CPD) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
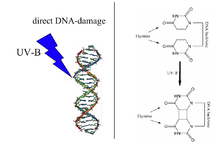 A UV radiation induced thymine-thymine cyclobutane dimer (right) is the type of DNA damage which is repaired by DNA photolyase. Note: The above diagram is incorrectly labelled as thymine as the structures lack 5-methyl groups. | |||||||||
| Identifiers | |||||||||
| EC no. | 4.1.99.3 | ||||||||
| CAS no. | 37290-70-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Photolyases (EC 4.1.99.3) are DNA repair enzymes that repair damage caused by exposure to ultraviolet light. These enzymes require visible light (from the violet/blue end of the spectrum) both for their own activation[1] and for the actual DNA repair.[2] The DNA repair mechanism involving photolyases is called photoreactivation. They mainly convert pyrimidine dimers into a normal pair of pyrimidine bases. Photo reactivation, the first DNA repair mechanism to be discovered, was described initially by Albert Kelner in 1949[3] and independently by Renato Dulbecco also in 1949.[4][5][6]
- ^ Yamamoto J, Shimizu K, Kanda T, Hosokawa Y, Iwai S, Plaza P, Müller P (October 2017). "Loss of Fourth Electron-Transferring Tryptophan in Animal (6-4) Photolyase Impairs DNA Repair Activity in Bacterial Cells". Biochemistry. 56 (40): 5356–64. doi:10.1021/acs.biochem.7b00366. PMID 28880077.
- ^ Thiagarajan V, Byrdin M, Eker AP, Müller P, Brettel K (June 2011). "Kinetics of cyclobutane thymine dimer splitting by DNA photolyase directly monitored in the UV". Proc Natl Acad Sci U S A. 108 (23): 9402–7. Bibcode:2011PNAS..108.9402T. doi:10.1073/pnas.1101026108. PMC 3111307. PMID 21606324.
- ^ Kelner A (February 1949). "Effect of Visible Light on the Recovery of Streptomyces Griseus Conidia from Ultra-violet Irradiation Injury". Proc Natl Acad Sci U S A. 35 (2): 73–9. doi:10.1073/pnas.35.2.73. PMC 1062964. PMID 16588862.
- ^ Dulbecco R (June 1949). "Reactivation of ultra-violet-inactivated bacteriophage by visible light". Nature. 163 (4155): 949. doi:10.1038/163949b0. PMID 18229246.
- ^ Dulbecco R (March 1950). "Experiments on photoreactivation of bacteriophages inactivated with ultraviolet radiation". J Bacteriol. 59 (3): 329–47. doi:10.1128/jb.59.3.329-347.1950. PMC 385765. PMID 15436402.
- ^ Friedberg EC (September 2015). "A history of the DNA repair and mutagenesis field: I. The discovery of enzymatic photoreactivation". DNA Repair (Amst). 33: 35–42. doi:10.1016/j.dnarep.2015.06.007. PMID 26151545.
© MMXXIII Rich X Search. We shall prevail. All rights reserved. Rich X Search